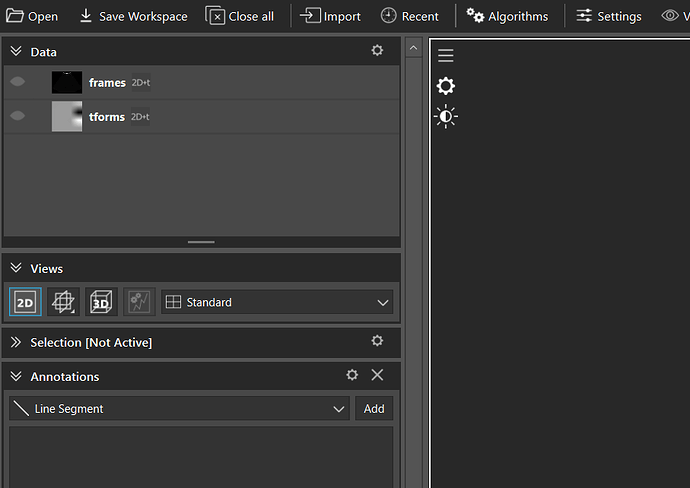
I have a HDF5 (.h5) file that contains two keys named “frames” and “tforms”. Basically contains 50 ultrasound images and their respective transformation matrix. When I import the file in ImFusion it shows the two sets of data, both as ‘2D+t’ images. How would I convert these two separate datasets to a ultrasound sweep so that it can be viewed as a volume ?
The HDF5 loader is a bit limited in terms of loading tracking, as you can see it will try to load as images.
You can try alternatives:
- Load the images from the HDF5 file, and load the tracking from a separate file, such as a .csv. Then you can join them together.
- Set the modality to Ultrasound using
SetModality - Select both images and tracking,
ConvertToUltrasound
- Set the modality to Ultrasound using
- Use the python console to load the HDF5 contents separately, as images and tracking, and follow the steps above.
For the python script something along the lines of:
import imfusion as imf
import h5py
import numpy as np
data = h5py.File(r"your_data_here.h5", "r")
images = np.array(data['frames']) # access your images using your keys
trackings = np.array(data['tforms']) # access your trackings using your keys
# Create a sweep from images and trackings
sweep = imf.UltrasoundSweep()
sweep_size = images.shape[0]
for img in images:
si = imf.SharedImage(img[:, :, np.newaxis])
sweep.add(si)
# if you have image timestamps use sweep.setTimestamp(timestamp, index)
tracking = imf.TrackingStream()
for pose in trackings:
tracking.add(pose) # use tracking.add(pose, timestamp) if you have tracking timestamps
sweep.addTracking(tracking)
Note that the demo version does not have access to the Ultrasound in python. If that’s the case, then just create a SharedImageSet and a TrackingStream, and you can merge them via the GUI